
Research
- Home
- Research
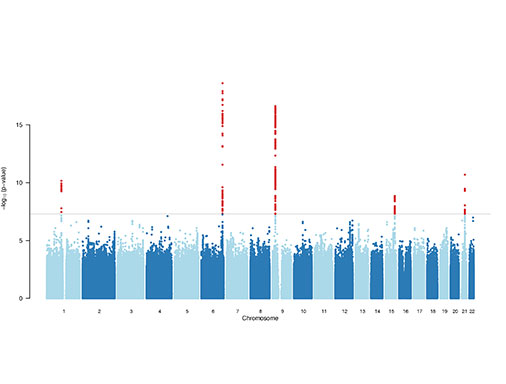
Map genetic variation and mechanisms associated with cardiovascular disease.
One of the enduring challenges of the post-GWAS and sequencing era is interpreting the function of non-coding variation, which often requires an understanding of how these variants regulate gene expression at the cellular and molecular levels. We previously employed genome-wide profiling of chromatin accessibility, transcription factor binding and gene expression in both naive and stimulated primary vascular cells and tissues. These molecular profiles have enabled identification of regulatory variants at candidate disease loci and elucidated potential mechanisms through transciption factor occupancy, nucleosome depletion, histone modification at enhancers, or altered miRNA binding. By integrating both expression and chromatin mapping with specific perturbations and functional assays we will focus on dissecting causal regulatory variants and mechanisms at other cardiovascular disease loci in the vessel wall.
Investigate role of genetic and environmental interactions in coronary artery disease.
Another poorly addressed issue in the field is the role of gene-gene (GxG) interactions (e.g. epistasis) and gene-environment (GxE) interactions in driving heritable risk for complex diseases. Our previous work at the TCF21 locus identified multiple physical interactions with other coronary artery disease (CAD) loci. To better understand biological epistasis in CAD, we are employing genome-wide screens for other disease-associated “master regulator” transcription factors in both normal and diseased states. These approaches will reveal insights into the combinatorial effects of genetic variation on the vascular wall interactome. We have also recently identified specific environmental factors interacting with smooth muscle CAD genes (e.g. TCF21) related to pro-inflammatory signaling. Both experimental and computational approaches will be employed to further explore these potential interactions across different individuals and disease states.
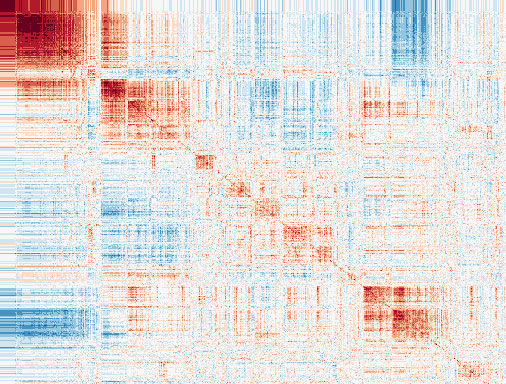
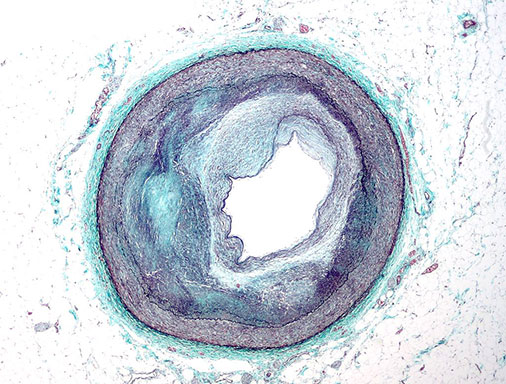
Investigate mural cell function and drug repositioning strategies for cardiovascular disease.
One of the distinguishing features of mural cells (smooth muscle cells and pericytes) is their ability to contract to regulate blood pressure and vascular tone. Aberrant contractile gene expression may lead to vascular dysfunction and hypertension, which is the leading heritable risk factor for cardiovascular disease. We have identified a number of contractile genes, which become dysregulated with atherosclerosis progression and have been associated with cardiovascular disease in human populations. We will investigate the impact of both non-coding and coding variation on SMC and pericyte function using high-throughput screening in human vascular tissue models under naïve and perturbed conditions. We will also employ a number of bioinformatics approaches to construct unbiased gene-target networks and explore repositioning opportunities as an individualized therapeutic approach.