Mokry M, Boltjes A, Cui K, […] Turner AW, Khan MD, Hodonsky CJ, […] Miller CL, Pasterkamp G. Transcriptomic-based clustering of human atherosclerotic plaques identifies subgroups with different underlying biology and clinical presentation. Nature Cardiovascular Research. 2022, December 12; 1140–1155.
Ma WF, Turner AW, Gancayco C, Wong D, Song Y, Mosquera JV, Auguste G, Hodonsky CJ, Prabhakar A, Ekiz HA, van der Laan SW, Miller CL. PlaqView 2.0: A comprehensive web portal for cardiovascular single-cell genomics. Frontiers in Cardiovascular Medicine. 2022, August 8; 9:969421.
Wang Y, Gao H, Wang F, […] Turner AW, […] Miller CL, Ross EG, Leeper NJ. Dynamic changes in chromatin accessibility are associated with the atherogenic transitioning of vascular smooth muscle cells. Cardiovascular Research. 2022,October 21; 118(13):2792-2804.
Ma L, Bryce NS, Turner AW, […] Miller CL, Björkegren JLM, Kovacic JC. The HDAC9-associated risk locus promotes coronary artery disease by governing TWIST1. PLoS Genetic. 2022, Jun2 17; 18(6):e1010261.
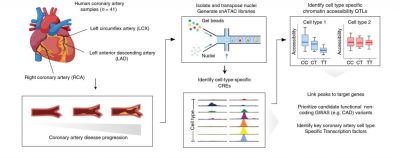
Turner AW, Hu SS, Mosquera JV, Ma WF, Hodonsky CJ, Wong D, Auguste G, Song Y, Sol-Church K, Farber E, Kundu S, Kundaje A, Lopez NG, Ma L, Ghosh SKB, Onengut-Gumuscu S, Ashley EA, Quertermous T, Finn AV, Leeper NJ, Kovacic JC, Björkgren JLM, Zang C, Miller CL. Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nature Genetics. 2022 June; 54(6):804-816.
Lino Cardenas CL, Jiang W, Cherbonneau F, […] Miller CL, Malhotra R. Novel treatment of small and large artery calcific disease via epigenetic activation of autophagy initiation genes. Research Square. 2022.05.03.
Slenders L, Landsmeer LPL, Cui K, […] Ma WF, Miller CL, […] Mokry M. Intersecting single-cell transcriptomics and genome-wide association studies identifies crucial cell populations and candidate genes for atherosclerosis. Eur Heart J Open. 2021 Dec 21;2(1):oeab043.
Ma WF, Hodonsky CJ, Turner AW, Wong D, Song Y, Mosquera JV, Ligay AV, Slenders L, Gancayco C, Pan H, Barrientos NB, Mai D, Alencar GF, Owsiany K, Owens GK, Reilly MP, Li M, Pasterkamp G, Mokry M, van der Laan SW, Khomtchouk BB, Miller CL. Enhanced single-cell RNA-seq workflow reveals coronary artery disease cellular cross-talk and candidate drug targets. Atherosclerosis. 2022, January 12, 340.
